We are developing a new version of Tumor Portal. You can go there and give us some feedback.
External References:
Wikipedia
GeneCards
HUGO
COSMIC
Google Scholar
NCBI Description of FAT1 |
| This gene is an ortholog of the Drosophila fat gene, which encodes a tumor suppressor essential for controlling cell proliferation during Drosophila development. The gene product is a member of the cadherin superfamily, a group of integral membrane proteins characterized by the presence of cadherin-type repeats. In addition to containing 34 tandem cadherin-type repeats, the gene product has five epidermal growth factor (EGF)-like repeats and one laminin A-G domain. This gene is expressed at high levels in a number of fetal epithelia. Its product probably functions as an adhesion molecule and/or signaling receptor, and is likely to be important in developmental processes and cell communication. Transcript variants derived from alternative splicing and/or alternative promoter usage exist, but they have not been fully described. |
Community Annotation of FAT1 Add / Edit FAT1: Annotations
No community annotations yet for FAT1.
|
|
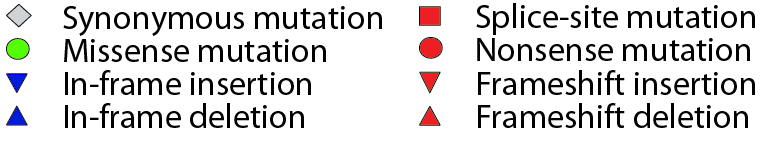
Figure notes
• "Mouse over" a mutation to see details. |
|
Click on a tumor type to see its full list of significant genes.
Data details